Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G51056AN
G51056AN
Summary
- GlyTouCan ID
-
G51056AN
- Motifs
- Glucosylceramide
3D Structures
GlycoShape
Organisms
| Organisms | Evidence |
|---|---|
| Leishmania mexicana | |
| Bradyrhizobium yuanmingense | |
| Bradyrhizobium elkanii | |
| Poria | |
| Agrobacterium sp. ATCC 31749 |
Taxonomic Hierarchy
Sequence Descriptors
- GlycoCT
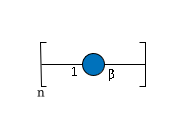
-
RES 1r:r1 REP REP1:2o(3+1)2d=-1--1 RES 2b:b-dglc-HEX-1:5
- WURCS
- WURCS=2.0/1,1,1/[a2122h-1b_1-5]/1/a1-a3~n
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 21665197 | Characterization of cyclic beta-glucans of Bradyrhizobium by MALDI-TOF mass spectrometry | Choma A | 2011 Sep 27 |
|
| 16310757 | Comparison of curdlan and its carboxymethylated derivative by means of Rheology, DSC, and AFM | Jin Y | 2006 Jan 16 |
|
| 8424790 | Purification and properties of three (1-->3)-beta-D-glucanase isoenzymes from young leaves of barley (Hordeum vulgare) | Hrmova M | 1993 Jan 15 |
|
| 8486553 | Isolation and characterization of a Bacillus strain capable of degrading the extracellular glucan from Cellulomonas flavigena strain KU | Bertram PA | 1993 Apr |
|
| 6400487 | Special bacterial polysaccharides and polysaccharases | Harada T | 1983 |
|
| 843521 | A 13C nuclear magnetic resonance study of gel-forming (1 goes to 3)-beta-d-glucans. Evidence of the presence of single-helical conformation in a resilient gel of a curdlan-type polysaccharide 13140 from Alcaligenes faecalis var. myxogenes IFO 13140 | Saito H | 1977 Mar 08 |
|