
GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE Glycan Converters
Glycan Converters
Glycan structure conversion tools between WURCS, GlycoCT, IUPAC, etc. Image generation and validation functionality are also available.
Glycan text formats
GlycoCT uses a similar graph concept to the KCF format (Herget et al. (2008)) and consists of two varieties: a condensed format and an XML format. The former allows for unique identification of glycan structures in a compact manner, while the latter facilitates data exchange. The monosaccharide namespace consists of five components and basically follows those defined by IUPAC: the basetype, anomeric configuration, the monosaccharide name with configurational prefix, chain length indicator, ring forming positions and further modification designators. Trivial names such as fucoso or rhamnoso are not permitted in GlycoCT.
The monosaccharide naming convention follows the following format: a-bccc-DDD-e:f|g:h, where a is the anomeric configuration (one of a, b, o, x), b is the stereoisorncr configuration (one of d, l, x), ccc is the three-letter code for the monosaccharide as listed in table below, DDD is the base type or superclass indicating the number of consecutive carbon atoms such as HEX, PEN, NON, e and f indicate the carbon numbers involved in closing the ring, g is the position of the modifier, and h is the type of modifier (one of d=deoxygenation, a=acidic function, keto=carbonyl function, en=double bond, aldi=reduction of C1-carbonyl, sp2=outgoing double bond linkage, sp=outgoing triple bond linkage, geminal=two identical substitutions). For a, b, e, f and g, an x can be used to specify an unknown value. bccc and g : h may also be repeated if necessary. Thus α-D-Galp would be represented as a-dgal-HEX-1:5 and α-D-Kdnp would be a-dgro-dgal-non-2:6,1a,2:keto,3:d in GlycoCT format.
It is noted that substituents of monosaccharides are also treated as separate residues attached to the base residue. These substituents are distinguished by specifying one of the following codes immediately after the residue number: b=basetype, s=substituent, r=repeating unit, a=alternative unit.
The GlycoCT format follows something similar to the KCF format, where the residues are specified in a RES section, and the linkages in a LIN section.
List of monosaccharide and their three-letter codes used in GlycoCT.
| Monosaccharide name | Three-letter code | Superclass |
|---|---|---|
| Allose | ALL | HEX |
| Altrose | ALT | HEX |
| Arabinose | ARA | PEN |
| Erythrose | ERY | TET |
| Galactose | GAL | HEX |
| Glucose | GLC | HEX |
| Glyceraldehyde | GRO | TRI |
| Gulose | GUL | HEX |
| Idose | IDO | HEX |
| Lyxose | LYX | PEN |
| Mannose | MAN | HEX |
| Ribose | RIB | PEN |
| Talose | TAL | HEX |
| Threose | TRE | TET |
| Xylose | XYL | PEN |
List of substituents used in GlycoCT.
| acetyl | amidino | amino |
| anhydro | bromo | chloro |
| diphospho | epoxy | ethanolamine |
| ethyl | fluoro | formyl |
| glycolyl | hydroxymethyl | imino |
| iodo | lactone | methyl |
| n-acetyl | n-alanine | n-amidino |
| n-dimethyl | n-formyl | n-glycolyl |
| n-methyl | n-methyl-carbomoyl | n-succinate |
| n-sulfate | n-triflouroacetyl | nitrate |
| phosphate | phospho-choline | phospho-ethanolamine |
| pyrophosphate | pyruvate | succinate |
| sulfate | thio | triphosphate |
| (r)-1-hydroxyethyl | (r)-carboxyethyl | (r)-carboxymethyl |
| (r)-lactate | (r)-pyruvate | (s)-1-hydroxyethyl |
| (s)-carboxyethyl | (s)-carboxymethyl | (s)-lactate |
| (s)-pyruvate | (x)-lactate | (x)-pyruvate |
Example of GlycoCT format: The N-glycan core structure represented in GlycoCT format.
RES
1b:b-dglc-HEX-1:5
2s:n-acetyl
3b:b-dglc-HEX-1:5
4s:n-acetyl
5b:b-dman-HEX-1:5
6b:a-dman-HEX-1:5
7b:a-dman-HEX-1:5
LIN
1:1d(2+1)2n
2:1o(4+1)3d
3:3d(2+1)4n
4:3o(4+1)5d
5:5o(3+1)6d
6:5o(6+1)7d
Although the two-dimensional notation of glycans as in figure below may be visually appealing, it is not suitable for storage in a database, and bioinformatic analysis tools would not be able to make use of it. Thus the IUPAC-IUBMB (International Union of Pure and Applied Chemistry - International Union of Biochemistry and Molecular Biology) has specified the "Nomenclature of Carbohydrates" to uniquely describe complex oligosaccharides based on a three-letter code to represent monosaccharides.
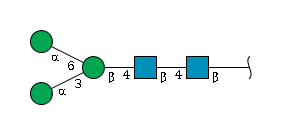
For example, gal represents galactose and man represents mannose; a listing of the common monosaccharides (and their derivatives) that occur in oligo- and polysaccharides is given in Table below (Tsai (2007)).
Each monosaccharide code is preceded by the anomeric descriptor and the configuration symbol. The ring size is indicated by an italic f for furanose or p for pyranose. The carbon numbers that link the two monosaccharide units are given in parentheses between the symbols separated by an arrow.
Double-headed arrows may be used if monosaccharides are linked through their anomeric centers. Moreover, α and β may be represented as a or b, respectively. In such a way, long carbohydrate sequences can be adequately described in abbreviated form using a sequence of letters.
Common monosaccharides (and their derivatives) that occur in oligo- and polysaccharides.
| Monosaccharide | IUPAC format |
|---|---|
| D-Ribose | D-Ribf |
| 2-Deoxy-D-Ribose | D-dRibf |
| D-Xylose | D-Xylf |
| L-Arabinose | L-Araf |
| D-Glucose | D-Glcp |
| D-Galactose | D-Galp |
| D-Mannose | D-Manp |
| D-Fructose | D-Fruf |
| L-Fucose | L-Fucp |
| L-Rhamnose | L-Rhap |
| D-Glucuronic acid | D-GlcpA |
| D-Galacturonic acid | D-GalpA |
| N-acetyl-D-glucosamine | D-GlcpNAc |
| N-acetyl-D-galactosamine | D-GalpNAc |
| N-acetylmuramic acid | D-MurpAc |
| N-acetylneuraminic acid | D-NeupAc |
| N-glycolylneuraminic acid | D-NeupGc |
Example of extended IUPAC format: The N-glycan core structure represented in extended IUPAC format.
α-D-Manp-(1→3)[α-D-Manp-(1→6)]-β-D-Manp-(1→4)-β-D-GlcpNAc-(1→4)-β-D-GlcpNAc-(1→
Example of condensed IUPAC format: The N-glycan core structure represented in condensed IUPAC format.
Man(a1-3)[Man(a1-6)]Man(b1-4)GlcNAc(b1-4)GlcNAc(b1-
Linear Code® is a carbohydrate format that uses a single-letter nomenclature for monosaccharides and includes a condensed description of the glycosidic linkages. Monosaccharide representation is based on the common structure of a monosaccharide where modifications to the common structure are indicated by specific symbols, as in the following (Banin et al. (2002)).
- Stereoisomers (D or L) differing from the common isomer are indicated by apostrophe (’).
- Monosaccharides with differing ring size (furanose or pyranose) from the common form are indicated by a caret (^).
- Monosaccharides differing in both of the above are indicated by a tilde (˜).
List of common monosaccharide structures and their single-letter code as used in the Linear Code® format. Note that all the sugars are assumed to be in pyranose form unless otherwise specified.
| Common configuration | Full name | Linear Code® |
|---|---|---|
| D-Glcp | D-Glucose | G |
| D-Galp | D-Galactose | A |
| D-GlcpNAc | N-Acetylglucosamine | GN |
| D-GalpNAc | N-Acetylgalactosamine | AN |
| D-Manp | D-Mannose | M |
| D-Neup5Ac | N-Acetylneuraminic acid | NN |
| D-Neup | Neuraminic acid | N |
| KDN | 2-Keto-3-deoxy-D-glycero-D-galacto-nonulosonic acid | K |
| Kdo | 3-deoxy-D-manno-2 Octulopyranosylono | W |
| D-GalpA | D-Galacturonic acid | L |
| D-ldop | D-loduronic acid | I |
| L-Rhap | L-Rhamnose | H |
| L-Fucp | L-Fucose | F |
| D-Xylp | D-Xylose | X |
| D-Ribp | D-Ribose | B |
| L-Araf | L-Arabinofuranose | R |
| D-GlcpA | D-Glucuronic acid | U |
| D-Allp | D-Allose | O |
| D-Apip | D-Apiose | P |
| D-Fruf | D-Fructofuranose | E |
List of common modifications as used in the Linear Code® format.
| Modification Type | Linear Code® |
|---|---|
| deacetylated N-acetyl | Q |
| ethanolaminephosphate | PE |
| inositol | IN |
| methyl | ME |
| N-acetyl | N |
| O-acetyl | T |
| phosphate | P |
| phosphocholine | PC |
| pyruvate | PYR |
| sulfate | S |
| sulfide | SH |
| 2-aminoethylphosphonic acid | EP |
Example of Linear Code®: The N-glycan core structure represented in Linear Code®.
Ma3(Ma6)Mb4GNb4GN
The KEGG Chemical Function (KCF) format for representing glycan structures was originally used to represent chemical structures (thus the name) in KEGG. KCF uses the graph notation, where nodes are monosaccharides and edges are glycosidic linkages. Thus to represent a glycan, at least three sections are required: ENTRY, NODE, EDGE, followed by three slashes ‘///’ at the end.
- The ENTRY section consists of one line and may specify a name for the structure followed by the keyword Glycan.
- The NODE section consists of several lines. The first line contains the number of monosaccharides or aglycon entities, and the following lines consist of the details of these entities numbered consecutively. For each entity line, the name and x- and y-coordinates (to draw on a 2D plane) must be specified.
- Similarly, the EDGE section consists of several lines, the first line containing the number of bonds (usually one less than the number of NODEs), followed by the details of the bond information. The format for the bond information is as follows:num <donor node#>:<anomeric configuration (a or b)><donor carbon#> <acceptor node#>:<acceptor carbon#>
Example of KCF format: The N-glycan core structure represented in KCF format.
ENTRY XYZ Glycan
NODE 5
1 GlcNAc 15.0 7.0
2 GlcNAc 8.0 7.0
3 Man 1.0 7.0
4 Man -6.0 12.0
5 Man -6.0 2.0
EDGE 4
1 2:b1 1:4
2 3:b1 2:4
3 5:a1 3:3
4 4:a1 3:6
///
Web3 Unique Representation of Carbohydrate Structures (WURCS) as a linear notation for representing carbohydrates for the Semantic Web.
Example of WURCS format: The N-glycan core structure represented in WURCS format.
WURCS=2.0/3,5,4/[a2122h-1b_1-5_2*NCC/3=O][a1122h-1b_1-5][a1122h-1a_1-5]/1-1-2-3-3/a4-b1_b4-c1_c3-d1_c6-e1
