Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G99225ZM
G99225ZM
Summary
- GlyTouCan ID
-
G99225ZM
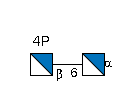
- IUPAC Condensed
- GlcN4P(b1-6)GlcN(a1-
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 420.11
External Links
- PubChem Compound
- PubChem Substance
- CarbBank
- 2366 2371 5269 7901 8070 8114 26154 26155 26156 26157 26158 26159 27372 30080 30081 31679 31916 32578 32579 32580 32619 32621 32622 33466 33467 34872 35293 36912 37554 37555 37893 37908 37914 37915 37917 38149 38219 38228 38230 39421 41174 41206 41208 41212 41214 41215 41216 41217 41218 41219 41220 41222 41223 42040 42921 42924 42966 44438 44468 44545 45370 45371 45372 45604 46047 46049 46289 46290 46292 46294 46296 46297 46299 46301 46303 46305 46306 46339 46340 46911 46912 46913 46914 46916 47337 47338 47340 47341 47342 47343 47344 47345 47346 47348 47349 47350 47353 47357 47358 47360 47362 47363 47365 47367 47533 47743 48486 49212 49214 49216 50433 50443 50647 50648 50658 50913
Organisms
| Organisms | Evidence |
|---|---|
| Vibrio cholerae | |
| Rubrivivax gelatinosus | |
| Neisseria meningitidis | |
| Escherichia coli | |
| Chromobacterium violaceum |
Taxonomic Hierarchy
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dglc-HEX-1:5 2s:amino 3b:b-dglc-HEX-1:5 4s:amino 5s:phosphate LIN 1:1d(2+1)2n 2:1o(6+1)3d 3:3d(2+1)4n 4:3o(4+1)5n
- WURCS
- WURCS=2.0/2,2,1/[a2122h-1a_1-5_2*N][a2122h-1b_1-5_2*N_4*OPO/3O/3=O]/1-2/a6-b1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 26085093 | Structural Basis for Antibody Recognition of Lipid A: INSIGHTS TO POLYSPECIFICITY TOWARD SINGLE-STRANDED DNA | Haji-Ghassemi O | 2015 Aug 07 |
|
| 8188347 | Molecular structures that influence the immunomodulatory properties of the lipid A and inner core region oligosaccharides of bacterial lipopolysaccharides | Baker PJ | 1994 Jun |
|
| 7942255 | Molecular structure of lipid A, the endotoxic center of bacterial lipopolysaccharides | Zähringer U | 1994 |
|
| 8330896 | The chemical structure of bacterial endotoxin in relation to bioactivity | Rietschel ET | 1993 Apr |
|
| 1904818 | Structural characterization of the lipid A component of Pseudomonas aeruginosa wild-type and rough mutant lipopolysaccharides | Kulshin VA | 1991 Jun 15 |
|