Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
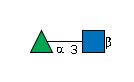
MIRAGE G99436AO
G99436AO
Summary
- GlyTouCan ID
-
G99436AO
- IUPAC Condensed
- Rha(a1-3)GlcNAc(b1-
- Subsumption Level
- Fully-defined saccharide
- Monoisotopic Mass
- 367.15
Organisms
| Organisms | Evidence |
|---|---|
| Shigella flexneri Y | |
| Shigella flexneri |
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O105 | Escherichia coli |
| E.coli O129 | Escherichia coli |
| E.coli O13 | Escherichia coli |
| E.coli O132 | Escherichia coli |
| E.coli O150 | Escherichia coli |
| E.coli O35 | Escherichia coli |
| E.coli O48 | Escherichia coli |
| E.coli O75 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:b-dglc-HEX-1:5 2s:n-acetyl 3b:a-lman-HEX-1:5|6:d LIN 1:1d(2+1)2n 2:1o(3+1)3d
- WURCS
- WURCS=2.0/2,2,1/[a2122h-1b_1-5_2*NCC/3=O][a2211m-1a_1-5]/1-2/a3-b1
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 15979596 | Structure of the O-polysaccharide of Proteus mirabilis OC (CCUG 10702) from a new proposed Proteus serogroup O75 | Zabłotni A | 2005 Aug 15 |
|