Table Filtering
Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / Organisms GlycoBase
GlycoBase
GlycoBase is a relational database which contains the HPLC elution positions for over 350 2-AB labelled N-glycan structures together with predicted products of exoglycosidase digestions.
GlycoBase and autoGU: tools for HPLC-based glycan analysis
Campbell MP, et al.
Bioinformatics. 2008 May 1;24(9):1214-6. PMID: 18344517
| Source | Last Updated |
|---|---|
| Glycobase | April 18, 2024 |
| GlyTouCan ID | GlycoBase ID | Compositions | Compositions (GlyTouCan ID) | Mass ▼ | GU (average) | GU (standard deviation) |
|---|---|---|---|---|---|---|
 G63069TR
G63069TR
|
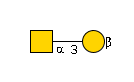
507 | Hexose1NeuAc1 |
 G76062CD
G76062CD
|
512.1644287109375 | 2.440000057220459 | 0.0 |
 G78074HX
G78074HX
|
1189 | Hexose3 |
 G99375FM
G99375FM
|
504.1690368652344 | 2.8715384006500244 | 34.458465576171875 |
 G45796AX
G45796AX
|
7088 | Hexose3 |
 G99375FM
G99375FM
|
504.1690368652344 | 2.9538462162017822 | 35.4461555480957 |
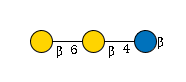 G60125IW
G60125IW
|
27862 | Hexose3 |
 G99375FM
G99375FM
|
504.1430969238281 | 3.1523077487945557 | 37.82769775390625 |
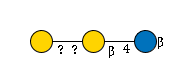 G82233EX
G82233EX
|
27866 | Hexose3 |
 G99375FM
G99375FM
|
504.1430969238281 | 3.299999952316284 | 6.600005149841309 |
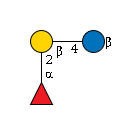 G61399NC
G61399NC
|
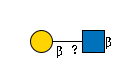
3909 | Fuc1Hexose2 | 488.17413330078125 | 2.386666774749756 | 19.09333610534668 | |
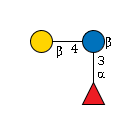 G57548NB
G57548NB
|
6289 | Fuc1Hexose2 | 488.17413330078125 | 2.502307653427124 | 30.027694702148438 | |
 G30207PZ
G30207PZ
|
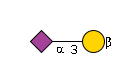
428 | Hexose1NeuAc1 |
 G76062CD
G76062CD
|
471.1588134765625 | 2.236666679382324 | 4.473410129547119 |
 G23447MZ
G23447MZ
|
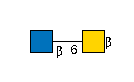
9572 | HexNAc2 |
 G71838YU
G71838YU
|
424.1693115234375 | 1.7949999570846558 | 1.7950069904327393 |
 G68491QD
G68491QD
|
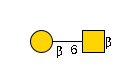
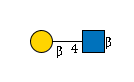
27235 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.14276123046875 | 2.0999999046325684 | 0.0 |
 G00055MO
G00055MO
|
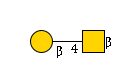
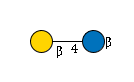
281 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.14276123046875 | 1.865384578704834 | 22.38461685180664 |
 G13003UU
G13003UU
|
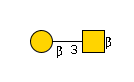
2898 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.14276123046875 | 2.0299999713897705 | 0.0 |
 G01534TU
G01534TU
|
3114 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.14276123046875 | 1.7921428680419922 | 23.297866821289062 |
 G60233WN
G60233WN
|
362 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.14276123046875 | 2.192500114440918 | 24.11798095703125 |
 G96642XC
G96642XC
|
27880 | HexNAc1Hexose1 |
 G29931IJ
G29931IJ
|
383.1224060058594 | 2.1600000858306885 | 10.800027847290039 |
 G84224TW
G84224TW
|
141 | Hexose2 |
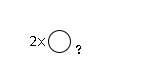 G07133YY
G07133YY
|
342.1162109375 | 1.9438461065292358 | 23.326156616210938 |
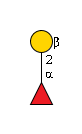 G00068MO
G00068MO
|
359 | Fuc1Hexose1 | 326.1213073730469 | 1.5750000476837158 | 1.5750079154968262 | |
 G61418DU
G61418DU
|
2529 | HexNAc1 |
 G70994MS
G70994MS
|
221.08993530273438 | 0.9049999713897705 | 2.7150046825408936 |
 G65889KE
G65889KE
|
135 | Hexose1 |
 G38776RF
G38776RF
|
180.06338500976562 | 1.01714289188385 | 13.222859382629395 |
