Table Filtering
Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE GlycoParaDB
GlycoParaDB
Glyco-enzyme reaction parameters estimated using Simulated Annealing via GlycoSim. The ranges of values for Km and Kmd that best matched experimental data are listed. The Parameter Estimation method has been described in this article.
| Enzyme | Gene ID | Uniprot ID | Substrate ID | BRENDA | Cazy family | Km_max | Km_min | Kmd_max | Kmd_min | taxonomyID | cell | Rhea | CHEBI |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta-1,4-galactosyltransferase 1 | 56336 | Q9JMK0 |
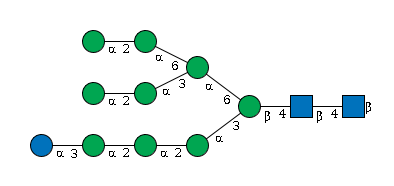 G19958IL
G19958IL
|
2.4.1.38 | GT7 |
|
|
|
|
10090 | ES cell | ||
| polypeptide N-acetylgalactosaminyltransferase | 14425 | P70419 |
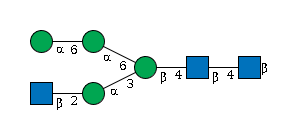 G75896PD
G75896PD
|
2.4.1.41 | GT27 |
|
|
|
|
10090 | ES cell | ||
| mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase | 17158 | P27046 |
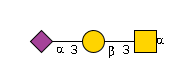 G65562ZE
G65562ZE
|
3.2.1.114 | GH38 |
|
|
|
|
10090 | ES cell | ||
| glycoprotein 6-alpha-L-fucosyltransferase | 53618 | Q9WTS2 |
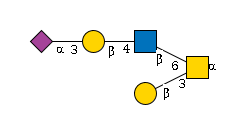 G11457RF
G11457RF
|
2.4.1.68 | GT23 |
|
|
|
|
10090 | ES cell | ||
| polypeptide N-acetylgalactosaminyltransferase | Q9JJ61 |
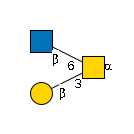 G00033MO
G00033MO
|
2.4.1.41 | GT27 |
|
|
|
|
10090 | ES cell | |||
| Beta-1,4-galactosyltransferase 1 | 14595 | P15535 |
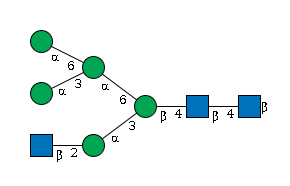 G08520NM
G08520NM
|
2.4.1.38 | GT7 |
|
|
|
|
10090 | ES cell | ||
| glycoprotein 6-alpha-L-fucosyltransferase | 53618 | Q9WTS2 |
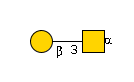 G00031MO
G00031MO
|
2.4.1.68 | GT23 |
|
|
|
|
10090 | ES cell | ||
| Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | 20446 | P70277 |
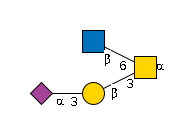 G85608AG
G85608AG
|
2.4.3.3 | GT29 |
|
|
|
|
10090 | ES cell | ||
| polypeptide N-acetylgalactosaminyltransferase | 270049 | E5D8G1 |
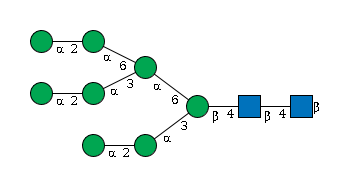 G89864BN
G89864BN
|
2.4.1.41 | GT27 |
|
|
|
|
10090 | ES cell | ||
| polypeptide N-acetylgalactosaminyltransferase | 71685 | Q8BVG5 |
 G00031MO
G00031MO
|
2.4.1.41 | GT27 |
|
|
|
|
10090 | ES cell |
