Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G68544GH
G68544GH
Summary
- GlyTouCan ID
-
G68544GH
- IUPAC Condensed
- GlcNAc(a1-
3D Structures
- GLYCAM
- DGlcpNAca1-OH
Organisms
| Organisms | Evidence |
|---|---|
| Salmonella enterica subsp. arizonae | |
| Salmonella | |
| Haliclona | |
| Amphimedon viridis | |
| Dictyostelium discoideum |
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Reference |
|---|---|---|---|---|
| B3GNT2 | UDP-Gal |
 G68544GH
G68544GH
|
[alpha]1-OBn | |
| B4GALT3 | UDP-Gal |
 G68544GH
G68544GH
|
o-Nitrophenyl-[alpha] | |
| B4GALT4 | UDP-Gal |
 G68544GH
G68544GH
|
[alpha]-1-Methyl | |
| B4GALT3 | UDP-Gal |
 G68544GH
G68544GH
|
Benzyl-[alpha] | |
| B4GALT2 | UDP-Gal |
 G68544GH
G68544GH
|
o-Nitrophenyl-[alpha] |
KEGG BRITE Database
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P04988 | Cysteine proteinase 1 | ||
| P14328 | Spore coat protein SP96 |
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O101 | Escherichia coli |
| E.coli O103 | Escherichia coli |
| E.coli O107 | Escherichia coli |
| E.coli O108 | Escherichia coli |
| E.coli O113 | Escherichia coli |
| E.coli O114 | Escherichia coli |
| E.coli O116 | Escherichia coli |
| E.coli O119 | Escherichia coli |
| E.coli O12 | Escherichia coli |
| E.coli O121 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-dglc-HEX-1:5 2s:n-acetyl LIN 1:1d(2+1)2n
- WURCS
- WURCS=2.0/1,1,0/[a2122h-1a_1-5_2*NCC/3=O]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 25945578 | Three-dimensional structure and ligand-binding site of carp fishelectin (FEL) | Capaldi S | 2015 Apr 24 |
|
| 25865483 | Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection | Wu X | 2015 Apr 23 |
|
| 25231460 | New structures and composition of cell wall teichoic acids from Nocardiopsis synnemataformans, Nocardiopsis\n halotolerans, Nocardiopsis composta and Nocardiopsis metallicus: a chemotaxonomic value | Tul'skaya EM | 2014 Sep 18 |
|
| 24915076 | GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) bound to three magnesium ions and ATP at the active site | Vithani N | 2014 May 10 |
|
| 24810719 | Frontispiece: Structural Basis of Chitin Oligosaccharide Deacetylation | Andrés E | 2014 May 08 |
|
| 24674022 | <scp>X</scp>‐ray structure of a novel endolysin encoded by episomal phage <scp>phiSM</scp>101 of <scp>C</scp>lostridium perfringens | Tamai E | 2014 Mar 05 |
|
| 24816108 | Insights into the binding specificity and catalytic mechanism ofN-acetylhexosamine 1-phosphate kinases through multiple reaction complexes | Wang KC | 2014 Apr 30 |
|
| 24065834 | Directed evolution to enhance thermostability of galacto-N-biose/lacto-N-biose I phosphorylase | Koyama Y | 2013 Sep 24 |
|
| 23485416 | Crystal Structures Identify an Atypical Two-Metal-Ion Mechanism for Uridyltransfer in GlmU: Its Significance to Sugar Nucleotidyl Transferases | Jagtap PKA | 2013 May 27 |
|
| 24217250 | Common Polymorphisms in Human Langerin Change Specificity for Glycan Ligands | Feinberg H | 2013 Dec 27 |
|
 G14843DJ
G14843DJ
 G49108TO
G49108TO
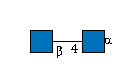 G05026ZL
G05026ZL
 G94923PG
G94923PG
