Submissions






GlyTouCan
Glycan Structure Repository
GlyComb
Glycoconjugate Repository
GlycoPOST
Glycomics MS raw data RepositoryUniCarb-DR
Glycomics MS Repository for glycan annotations from GlycoWorkbench
LM-GlycoRepo
Repository for lectin-assisted multimodality dataAll Resources
Genes / Proteins / Lipids Glycans / Glycoconjugates Glycomes Pathways / Interactions / Diseases / OrganismsTools
Guidelines
MIRAGE G96881BQ
G96881BQ
3D Structures
- GLYCAM
- LFucpa1-OH
Organisms
Taxonomic Hierarchy
GlycoGene Database (GGDB)
| Gene Symbol | Donor | Acceptor | Reducing terminal(Acceptor) | Product | Reducing terminal(Product) | Reference |
|---|---|---|---|---|---|---|
| B3GLCT | UDP-Glc |
 G96881BQ
G96881BQ
|
rat F-spondin TSR4 |
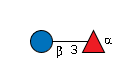 G36855WW
G36855WW
|
rat F-spondin TSR4 | |
| B3GLCT | UDP-Glc |
 G96881BQ
G96881BQ
|
[alpha]-pNp |
 G36855WW
G36855WW
|
[alpha]-pNp | |
| MFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
-pNP |
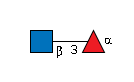 G26238GL
G26238GL
|
-pNP | |
| LFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
[alpha]-pNp |
 G26238GL
G26238GL
|
[alpha]-pNp | |
| LFNG | UDP-GlcNAc |
 G96881BQ
G96881BQ
|
-O-EGF |
 G26238GL
G26238GL
|
-O-EGF |
KEGG BRITE Database
- KEGG ID
Core Protein
| UniProt ID | Protein Name | Reference | Source |
|---|---|---|---|
| P00740 | Coagulation factor IX | ||
| P00749 | Urokinase-type plasminogen activator | ||
| P00750 | Tissue-type plasminogen activator | ||
| P07996 | Thrombospondin-1 | ||
| P08709 | Coagulation factor VII | ||
| P14328 | Spore coat protein SP96 | ||
| Q09163 | Protein delta homolog 1 | ||
| Q76LX8 | A disintegrin and metalloproteinase with thrombospondin motifs 13 |
Pathway
| Pathway Name | Organism |
|---|---|
| E.coli O11 | Escherichia coli |
| E.coli O125ab | Escherichia coli |
| E.coli O126H27 | Escherichia coli |
| E.coli O127 | Escherichia coli |
| E.coli O128ab | Escherichia coli |
| E.coli O128ab-H27 | Escherichia coli |
| E.coli O128ac | Escherichia coli |
| E.coli O156 | Escherichia coli |
| E.coli O157 | Escherichia coli |
| E.coli O159 | Escherichia coli |
Sequence Descriptors
- GlycoCT
-
RES 1b:a-lgal-HEX-1:5|6:d
- WURCS
- WURCS=2.0/1,1,0/[a1221m-1a_1-5]/1/
Literature
| PubMed ID | Title | First Author | Publication Date | Source |
|---|---|---|---|---|
| 30108726 | Triazolopyrimidines identified as reversible myeloperoxidase inhibitors | Duclos F | 2017 Nov 01 |
|
| 28254785 | Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity | Luca VC | 2017 Mar 24 |
|
| 28331984 | Structure of the N-glycans from the chlorovirus NE-JV-1 | Speciale I | 2017 Mar 22 |
|
| 29232414 | Influence of Trp flipping on carbohydrate binding in lectins. An example on Aleuria aurantia lectin AAL | Houser J | 2017 Dec 12 |
|
| 28806750 | Characterization of novel bangle lectin from Photorhabdus asymbiotica with dual sugar-binding specificity and its effect on host immunity | Jančaříková G | 2017 Aug 14 |
|
| 28572448 | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands | Suckling RJ | 2017 Aug 01 |
|
| 28393287 | Starch-enriched diet modulates the glucidic profile in the rat colonic mucosa | Gabrielli MG | 2017 Apr 09 |
|
| 27756822 | Dirigent Protein Mode of Action Revealed by the Crystal Structure of AtDIR6 | Gasper R | 2016 Oct 17 |
|
| 27758853 | A Novel Fucose-binding Lectin from Photorhabdus luminescens (PLL) with an Unusual Heptabladed β-Propeller Tetrameric Structure | Kumar A | 2016 Nov 25 |
|
| 27858203 | Enhanced fucosylation of GA1 in the digestive tracts of X-ray-irradiated mice | Iwamori M | 2016 Nov 18 |
|
 G49112ZN
G49112ZN
 G85196CF
G85196CF
